One powerful use case, yet often overlooked, of the autoencoders is anomaly detection.
In this tutorial, I will show how to use autoencoders to detect abnormal electrocardiograms (ECG).
I will divide the tutorial in two parts. The first one, here will guide you throught the problem formulation and how to train the autoencoder neural network over the ECG data.
I will release the next part in a week or two, so please subscribe to have the update!
What is anomaly detection?
A customer using an ATM, an electrocardiogram measured from a patient in a hospital, a connection status to a server, a machine operating on some task, etc. These may all be described as situations with two clearly defined statuses.
- A correct status, where things work as expected.
- An abnormal status, which would be the other case.
In the case of a customer using an ATM, an abnormal status would be caused by the user being an attacker, or by the system not working properly.
It is crucial to automatically detect such abnormal situations, which will save security or maintenance cost.
Thanks to their ability to condense data into a meaningful representation, autoencoders can be used to detect abnormal behaviors.
Autoencoders
Autoencoders have been tremendously useful for denoising data and data compression.
These models are able to compress input data into a meaningful internal representation, that can be reused for downstream tasks, such as text summarization, machine translation, object detection in images, and so on.
Their versatility makes their strength. They have been first presented in 1986 using multi-layer perceptrons as their building blocks, but have been shown to adapt quite well to many types of data, by changing their architectures. For example, they were used to improve the results of image classification, by basing their architecture on convolutional neural networks.
Their architecture is quite simple. One part, called the encoder, is responsible for extracting an internal representation from input data. The other part, called the decoder, is responsible for reconstructing data back to its original form, given the internal representation.
How to use autoencoders for anomaly detection?
Remember that autoencoders are good at one thing, which is to extract a meaningful representation out of some data. This might sound esoteric, but let's explain why it will help in anomaly detection.
An autoencoder is trained using unsupervised learning, on some unlabeled data, to reconstruct its input data, after having calculated an internal representation of it. By doing so, the autoencoder gets better at optimizing some criterion on the "normal data" subset we can gather, from all the possible input data.
By definition, abnormal data should show some peculiar features, which would make the autoencoder's task of reconstructing the abnormal data from the internal representation harder. But this is exactly what we want!
At this point, we can define a protocol for us tackle anomaly detection using autoencoders.
- Train an autoencoder on our normal data subset.
- Compare the autoencoder's criterion on the normal data subset, and the abnormal data subset.
- Define a threshold for us to safely classify data as abnormal or normal.
This last step is trickier than it sounds, as it will highly depend on our understanding of the problem and whether we are using the right tools.
Quick exploratory data analysis of electrocardiogram data
The ECG5000 dataset http://www.timeseriesclassification.com/description.php?Dataset=ECG5000 is a collection of time-series data of measured heartbeat from patients with severe congestive heart failure. The data is labelled by 0 if the heartbeat is abnormal, and 1 if it is normal.
Let's get the data from this Tensorflow tutorial.
dataframe = pd.read_csv('http://storage.googleapis.com/download.tensorflow.org/data/ecg.csv', header=None)
raw_data = dataframe.values
dataframe.head()We can see that the labels (0 or 1 values) of each ECG is on the last column, and the other columns are the time-series data points of the ECG.
labels = raw_data[:, -1]
data = raw_data[:, 0:-1]Let's visualize some data. The following will plots two samples, one normal, one abnormal, that I randomly chose from the dataset.
normal_ecg = data[labels == 1][16]
abnormal_ecg = data[labels == 0][5]
import matplotlib.pyplot as plt
plt.plot(normal_ecg, label="Normal")
plt.plot(abnormal_ecg, label="Abnormal")
plt.legend()
plt.show()
One sample of normal and abnormal data.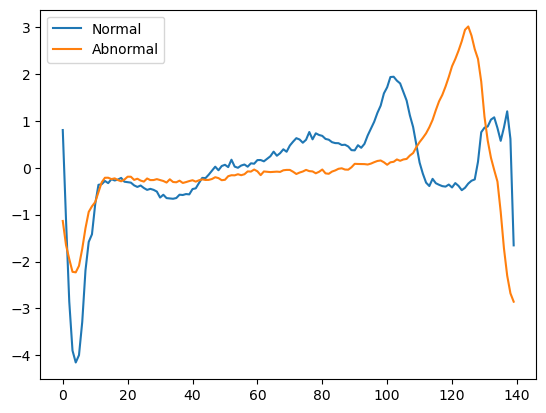
We can see some differences between the two curves, but one sample comparison does not give much information on the general patterns that a normal ECG should follow. Let's go further and plot the mean ECG for both our normal and abnormal samples.
normal_ecg_mean = data[labels == 1].mean(axis=0)
abnormal_ecg_mean = data[labels == 0].mean(axis=0)
import matplotlib.pyplot as plt
plt.plot(normal_ecg_mean, label="Normal mean")
plt.plot(abnormal_ecg_mean, label="Abnormal mean")
plt.legend()
plt.show()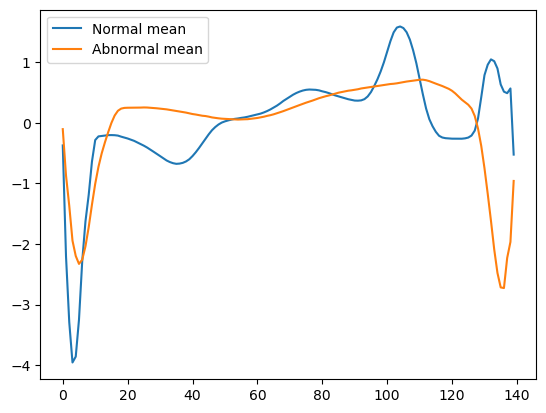
Normal and abnormal data mean.
Interestingly, we can note some striking differences.
- Normal data tend to have a lower minimum than abnormal data, appearing at around the first timesteps.
- Normal data tend to have a maximum value occurring at around timestep 100. This is not the case for abnormal data.
- Normal data tend to have a second spike near the end of the ECG which is not found on abnormal data.
We now seem to have quite a fair amount of information to classify data as "normal" or "abnormal", but how to transfer this intuitive knowledge into some reliable heuristic? And would our quick analysis of the mean values suffice to get good generalization?
It may be tempting to start hand-crafting some criterion for the classification problem but a better approach is to use machine learning in this case, because of the complexity of the problem.
Build an autoencoder for time-series data
Due to the relatively small size of the ECG samples (only 140 values per data sample), we can use an autoencoder architecture formed of multi-layer perceptrons.
import torch
class Autoencoder(torch.nn.Module):
def __init__(self, input_size=140):
super(Autoencoder, self).__init__()
self.enc1 = torch.nn.Linear(input_size, 32)
self.enc2 = torch.nn.Linear(32, 16)
self.enc3 = torch.nn.Linear(16, 8)
self.relu = torch.nn.functional.relu
self.sigmoid = torch.sigmoid
self.dec1 = torch.nn.Linear(8, 16)
self.dec2 = torch.nn.Linear(16, 32)
self.dec3 = torch.nn.Linear(32, input_size)
def encoder(self, x):
outputs = self.enc1(x)
outputs = self.relu(outputs)
outputs = self.enc2(outputs)
outputs = self.relu(outputs)
outputs = self.enc3(outputs)
outputs = self.relu(outputs)
return outputs
def decoder(self, x):
outputs = self.dec1(x)
outputs = self.relu(outputs)
outputs = self.dec2(outputs)
outputs = self.relu(outputs)
outputs = self.dec3(outputs)
outputs = self.sigmoid(outputs)
return outputs
def forward(self, x):
encoded = self.encoder(x)
decoded = self.decoder(encoded)
return decoded
autoencoder = Autoencoder()The reasons behind using a particular architecture are often empirically based on past research, and we can note the following.
- ReLU activation layers are used to improve the autoencoder's ability to learn non-linear representations.
- Using a stack of linear layers gives more depth to the network, and has been shown to provide faster convergence. This is one of the results behind the "deep" in Deep learning.
- The encoder progressively compresses the input data into an internal representation with smaller size (8-dimensional vector). This is to force the autoencoder to learn meaningful representation of the data, by discarding unnecessary information, while optimizing the criterion we will introduce in the following.
To train our autoencoder we need two things.
- A criterion, or loss function.
- An optimization strategy.
For time-series data, it is very common and wise to use some loss function based on the residuals (the error values obtained by subtracting the target values with the outputs values, for each timestep.). We will use the mean absolute error, or L1 loss, in PyTorch. Finally, let's use Adam optimizer, one of the most robust optimizers in current research.
optimizer = torch.optim.Adam(autoencoder.parameters())
criterion = torch.nn.L1Loss()We can keep only the normal data as training data. But to give us more confidence on the final result, let's split our training data into a train and test split. The test split won't be used for training the autoencoder, but will be used to test it against unseen data and have an idea of its ability to generalize to unseen data, that is, to correctly classify as normal data, normal ECG data it has never seen before.
import random
def train_test_split(data, labels, test_size=0.2, random_state=None):
"""Split the data into train and test sets."""
# Decide the split index
split_index = int(len(data) * test_size)
# Randomly shuffle the indices
random.seed(a=random_state)
random_indices = list(range(len(data)))
random.shuffle(random_indices)
# Shuffle the data
shuffled_data = data[random_indices]
shuffled_labels = labels[random_indices]
# Split the data
test_data, train_data = shuffled_data[:split_index], shuffled_data[split_index:]
test_labels, train_labels = shuffled_labels[:split_index], shuffled_labels[split_index:]
return train_data, test_data, train_labels, test_labelsLet's compute our train and test split. We also normalize the data for the autoencoder. Although anomalous data is not used during training, we keep it in the data when we split in train and test sets, to be able to make correct assumptions over the data distribution.
import torch
# Split the data
train_data, test_data, train_labels, test_labels = train_test_split(
data, labels, test_size=0.2, random_state=21
)
# Normalize the data
min_val = train_data.min()
max_val = train_data.max()
train_data = (train_data - min_val) / (max_val - min_val)
test_data = (test_data - min_val) / (max_val - min_val)
train_data = torch.tensor(train_data, dtype=float)
test_data = torch.tensor(test_data, dtype=float)
train_labels = train_labels.astype(bool)
test_labels = test_labels.astype(bool)
normal_train_data = train_data[train_labels]
normal_test_data = test_data[test_labels]
anomalous_train_data = train_data[~train_labels]
anomalous_test_data = test_data[~test_labels]With everything arranged, we can now run the training of the autoencoder.
from tqdm import trange
batch_size = 32
epochs = 100
losses = []
autoencoder = autoencoder.train()
autoencoder = autoencoder.requires_grad_(True)
n_batches = len(train_data) // batch_size
with trange(epochs) as tbar:
for epoch in tbar:
epoch_loss = 0.
for i in range(0, len(normal_train_data), batch_size):
batch = normal_train_data[i:i+batch_size]
optimizer.zero_grad()
outputs = autoencoder(batch.float())
loss = criterion(outputs, batch.float())
epoch_loss += loss.item()
loss.backward()
optimizer.step()
losses.append(epoch_loss)
tbar.set_postfix(loss=epoch_loss / float(n_batches))And see the learning curve.
Autoencoder training loss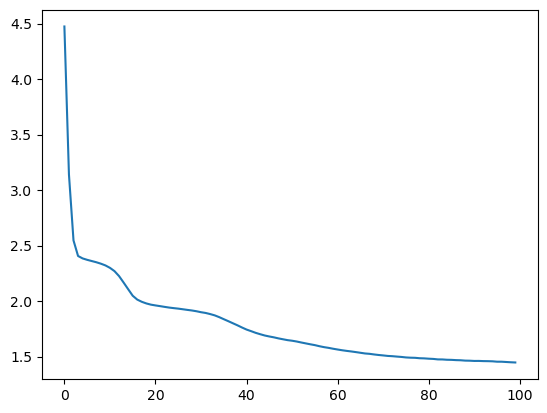
This looks good! The loss decreased to lower magnitudes of values. Let's go ahead and reconstruct some data. We can use the normal and abnormal ECG data above.
Conclusion
We saw how we can train an autoencodder over electrocardiogram (ECG) data. In my following article, I will show how to use our trained autoencoder to actually detect anomalies using a carefully tuned threshold.
Please subscribe to see my upcoming articles!